Raman spectroscopy is a scattering technique that
relies on the inelastic scattering of photons. The excitation of molecular bonds results in the shifts of the energy of
laser photons and thus frequency change of the incident light. Therefore, a
compound can have a specific Raman spectrum, while a Single-cell Raman Spectrum
(SCRS) can depict the overall profile of metabolites in the cell, i.e., the
metabolite state of the cell at a particular instance.
We have proposed the "Ramanome" concept, which is the collection of the SCRS from a cellular population (or a consortium, for “Meta-ramanome”) at a given instance. Each SCRS can have over 1500 Raman peaks, while each peak or combination of peaks can potentially represent a metabolic phenotype, therefore a ramanome data point would capture the single-cell-resolution metabolic phenome of the system at a certain state. Notably, within a ramanome, even though the genomes of the cells are identical, the SCRS of the cells are distinct; such intercellular heterogeneity of SCRS is an inherent feature of cellular systems, and is critical for single-cell analysis and interpretion.

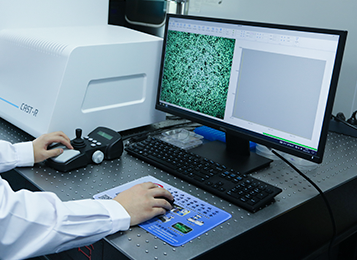
Ramanome has broad applications. For example, to meet the challenge in precision medicine, we invented the D2O-Ramanometry technology to rapidly measure stress response at single-cell level, and proposed the concept of MIC-MA (the lowest drug dose at which the metabolic activity of "every" cell is "completely" inhibited). As an innovative parameter to quantify drug sensitivity, MIC-MA is faster, more accurate, deeper in revealing mechanism, broader in application and easier in measurement than the traditional MIC. Then, we have developed the first Clinical Antimicrobial Susceptibility Test Ramanometry (CAST-R), and established the "Clinical Demonstration Network for Rapid AST at Single-cell level" together with clinical partners throughout China. MIC-MA/CAST-R has demonstrated its advantages in the treatment of acute bloodstream infections (tigecycline, etc.), chronic infections (Helicobacter pylori, etc.), oral disinfectant efficacy evaluation and screening (for bacteria and fungi), and highly sensitive drug resistance phenotyping of mutants. This principle is also applicable to the evaluation and screening of metabolic activity and/or resistance for industrial/environmental microorganisms.

In order to obtain the single cells of
targeted metabolic phenome from a cellular population or consortium (for e.g., single-cell
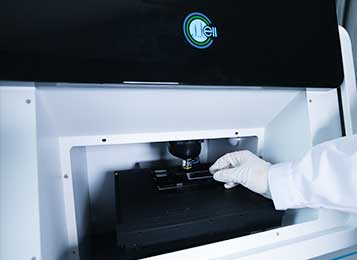
sequencing or culture), we have developed and commercialized a series of Raman-guided
single-cell sorting devices. These Raman-activated Cell Sorting (RACS) technologies are broadly classified into two
categories: stationary and microflow, depending on the motion state of cells at
the time of Raman acquisition.
The former includes Raman tweezers, Raman-Activated Cell Ejection (RACE and its upgraded version), and Raman-Activated Gravity-driven single-cell Encapsulation (RAGE), etc. This category of RACS devices mainly focuses on precise and indexed Raman-based single-cell collection and sorting. The latter, capable of sorting cells in a high-throughput manner, includes Raman-activated Microfluidic Sorting (RAMS), Raman-activated Droplet Sorting (RADS), and positive dielectrophoresis based Raman-activated Droplet Sorting (pDEP-RADS). In addition, we have introduced a number of downstream single-cell sequencing or culture technologies that are directly coupled to RACS, such as precisely-one-bacterial-cell high-coverage genome sequencing (RAGE-Seq), single-cell culture (RACS-Culture), and Addressable Dynamic Droplet Array (aDDA)-based single-cell sequencing technologies.
For Ramanome data analysis, we have proposed novel algorithms and concepts, such as Intra-Ramanome Correlation Analysis (IRCA) and Raman Barcode of Cellular-response to Stresses (RBCS). We also developed the Ramanome Ensemble Learning algorithm and published the first Micro-Algal Ramanome Database (MARD). These algorithms, software and databases are all generally applicable.
With the generous support of the National Natural Science Foundation of China, Ministry of Science and Technology of China, and Chinese Academy of Sciences, we have developed a series of Ramanome/RACS-based instruments, which have entered the market, such as Clinical Antimicrobial Susceptibility Test Ramanometry (CAST-R), Flow-mode Raman-activated Cell Sorter (FlowRACS), Raman-activated Single-Cell Sorting and Sequencing (RACS-Seq), Single-cell Microdroplet Sorting System (EasySort Lego / Compact), etc.
Using these original instruments, we have established a platform to support the entire workflow that links single-cell metabolic phenome profiling and the corresponding high-quality single-cell genome sequencing. Moreover, our instruments have been employed in many research areas, such as high-throughput screening of enzyme activity in vivo, rapid profiling of metabolic functions in cellular factories, high-throughput screening of metabolic phenotypes of mutants (oil-producing super microalgae induced by blue light), and reconstruction of metabolite-conversion networks. For microbiome samples, using RACS-Seq which is equipped with the RAGE chip, we have, for the first time, demonstrated the ability to simultaneously obtain metabolic phenome and high-coverage genome from precisely one bacterial cell, from both clinic samples (e.g., urine, gastric mucosa, etc.) and complex environmental samples (e.g., soil, ocean, etc.).
In addition, we established a new technique called single-cell Raman-activated Cell Sorting and Cultivation (scRACS-Culture), for metabolic-function based assessment and mining of live probiotics directly from complex environmental samples (such as mining in situ phosphate solubilizing microbes from urban wastewater), which are advantageous because it is of single-cell precision, culture-free, and does not require fluorescence probes. This "screen first, culture second" approach is a new route for functional mining of microbiomes.

By quantitatively converting the molecular spectrum of intracellular metabolites into key metabolic phenotypes such as substrate intake, product synthesis, response to environment and metabolite conversion, ramanome is a kind of “omics” that is closest to “function”, and a more direct way to probe the metabolism of cells. Moreover, ramanome-based tools offer many advantages, such as label-freeness, non-destructiveness, landscape-like phenotyping, high-speed, low-cost, automation, general applicability to all cell types, and ability to couple to downstream single-cell sequencing, mass spectrometry or cell culture. Thus ramanome is highly complementary to those existing omics tools, and together they formulate a complete single-cell multi-omics platform.
In summary,Ramanome Technology Platform (RTP), which establishes the link between metabolic phenome and genome (or transcriptome, proteome, etc) at the precision of one cell (or one organelle for an eukaryotic cell), answers the fundamental question of "Who is doing What, and How" at the deepest level. Therefore, RTP has become a powerful tool for functional mining and mechanistic dissection of human, animal, plant and microbial single-cells, and is serving a wide range of fields such as precision medicine, synthetic biology, one-health, biosecurity, and ecological surveillance.












 鲁公网安备37021202001515
鲁公网安备37021202001515


