RACS-Seq Raman-activated Optical Tweezers-based Cell Sorter
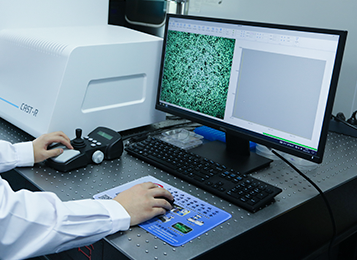
RACS-Seq® is an integrated instrument system that links single-cell metabolic phenome profiling, Raman-activated cell sorting, sample preparation for single-cell genome sequencing, and single-cell cultivation. Based on Single-cell Raman spectroscopy, RACS-Seq® allows direct classification of microbial species and measurement of a wide range of metabolic phenotypes (and their intercellular heterogeneity) for individual cells in a culture-free manner.
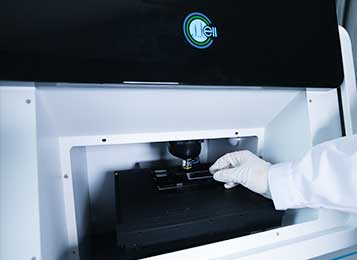
Moreover, in RACS-Seq®, each single-cell that is sorted based on its metabolic phenome can be encapsulated in a microdroplet for single-cell DNA/RNA amplification, enabling construction of high-coverage, low-bias single-cell nucleic acid libraries. In addition, single bacteria cell can be sorted and cultured in micro-droplets using this instrument.
The first commercialized instrument for Raman-activated single-cell sorting and sequencing; Funded by the Scientific Instrument Innovation Program of CAS; Funded by the Scientific Instrument Development Program of NSFC.
Contact Us